Bioprocessing Blog
Insights from digital bioprocessing experts
The Pursuit of Process Understanding
Quality by Design (QbD) transformed process development from a system that relied on end-product testing with little knowledge of the underlying process to one that made process understanding and risk management the focus. Under QbD, scientists are asked to understand how critical process parameters (CPPs) impact product quality and process productivity, and to embed robustness into the process itself.
Given the complexity of bioprocessing and the gaps in scientific understanding, scientists cannot rely exclusively on mechanistic models to capture all process dynamics and meet the objectives of QbD. In practice, depending solely on such models, particularly in complex operations like cultivation, leads to knowledge gaps and exposes both product and process to excessive risk
In contrast, pure machine learning models aim to uncover patterns directly from data without relying on prior knowledge. While AI’s impressive analytical capabilities have fueled adoption across many domains, applying these models to bioprocessing exposes critical limitations. The challenge lies in a fundamental data dichotomy: ML models typically demand large datasets for reliable performance, yet bioprocessing is inherently data-conservative, with experimental data generation being both costly and time-intensive.
As a result, simple statistical approaches such as linear regression have long been the go-to analytical technology for understanding the relationships between process parameters and performance, but not without limitations. Their linear structure oversimplifies the complex, non-linear behavior of bioprocesses, and they fail to capture process evolution or dynamics over time, offering only an end-point perspective. Although statistical approaches have long served as the industry standard, their oversimplification of complex systems underscores the need for more advanced methods capable of capturing the true nature of bioprocesses.

Experimental Design Dictated by Analytical Technologies
Experimental design strategies are often closely linked to the analytical tools employed. For example, One-Factor-At-a-Time (OFAT) approaches are commonly paired with simple visualization techniques, providing an intuitive but limited means of exploring process effects.
Statistical DoE that perturbs only the corner and center points of the design space overlooks the complex combinatorial interactions that emerge when multiple process parameters vary simultaneously. While such a design may suffice for fitting a linear regression, it inherently biases the representation of parameter–performance relationships in systems that are intrinsically non-linear. In bioprocesses, where dynamics and interdependencies govern outcomes, this approach risks oversimplification, masking critical effects within the interior of the design space and leading to incomplete or even misleading process understanding.
In addition, as the number of design factors increases, statistical DoE becomes inefficient, with experimental runs rising exponentially and full-factorial designs quickly becoming impractical. To reduce workload, practitioners use screening campaigns and focus only on selected factors or interactions. While this lowers the experimental burden, it risks missing higher-order or non-linear effects, leading to incomplete process understanding and additional risk.
Hybrid Models: a Paradigm Shift for Bioprocess Analytics
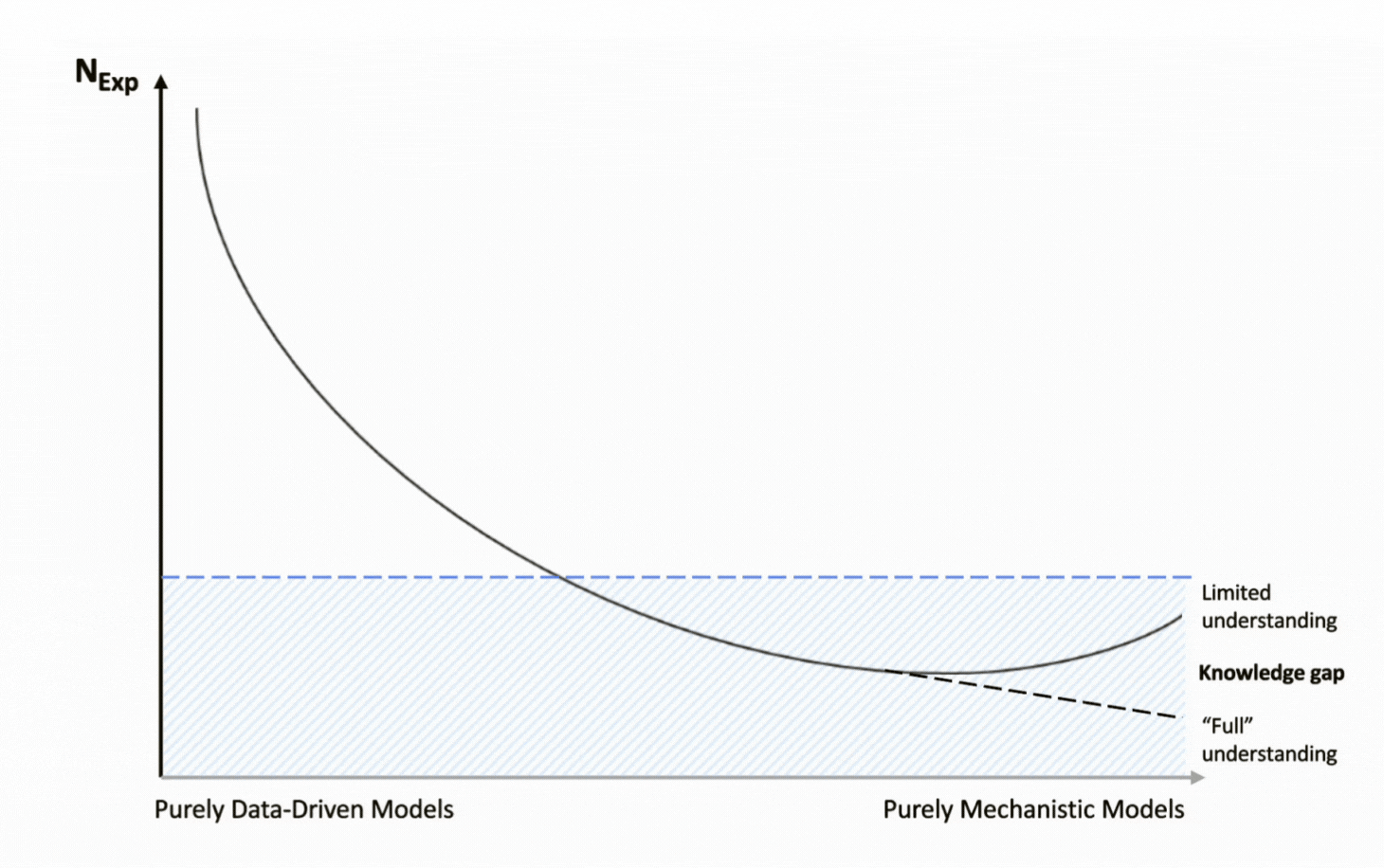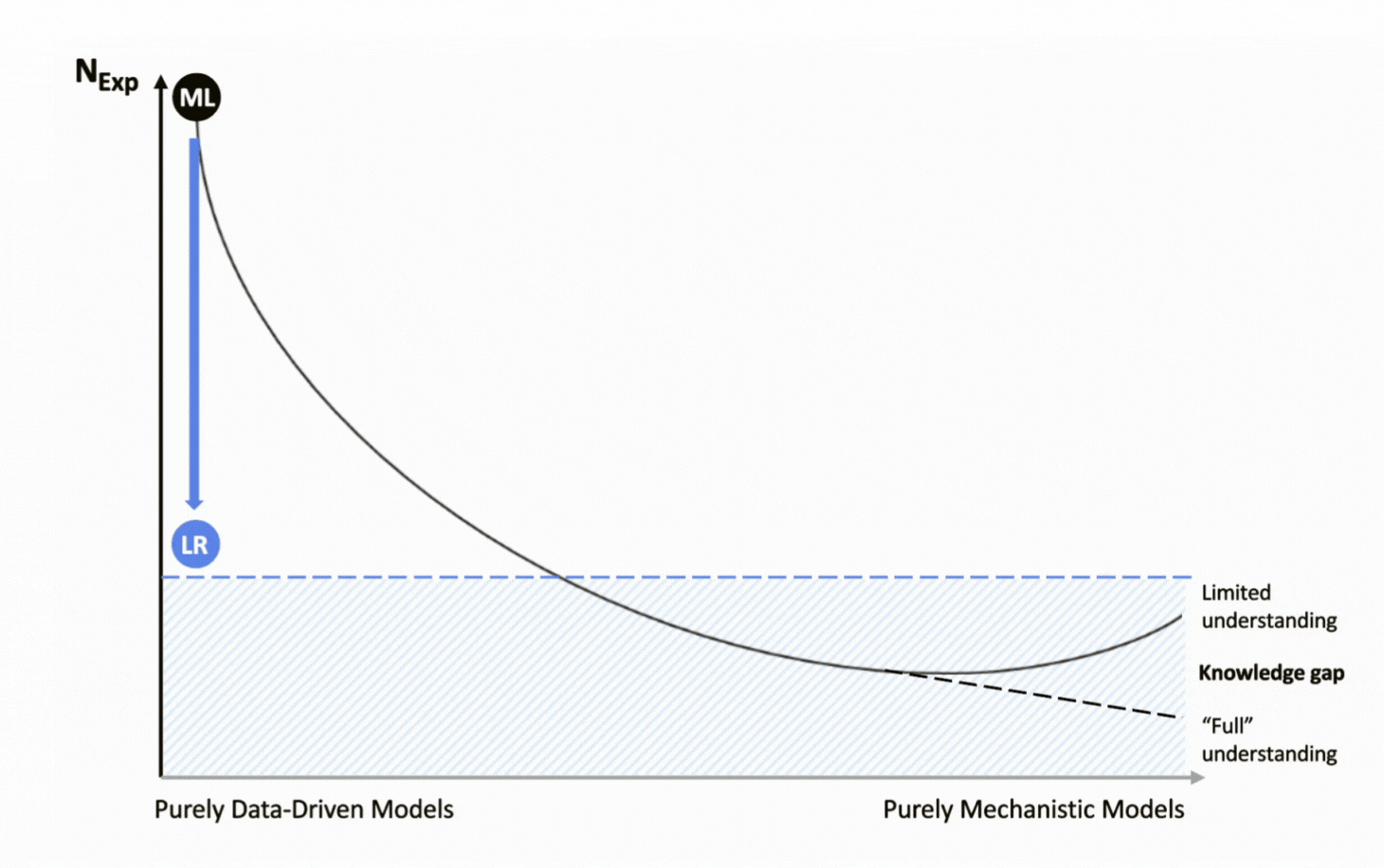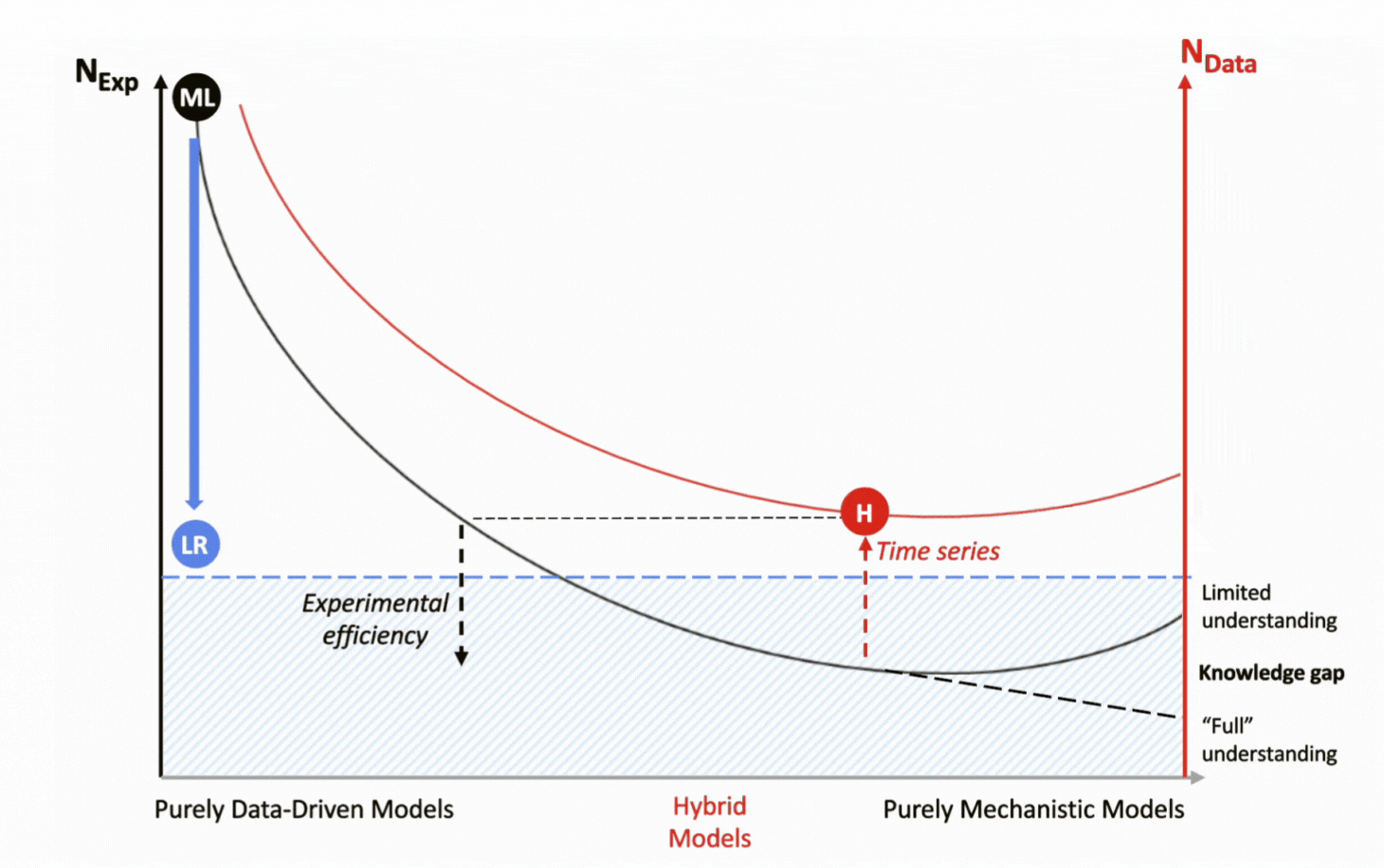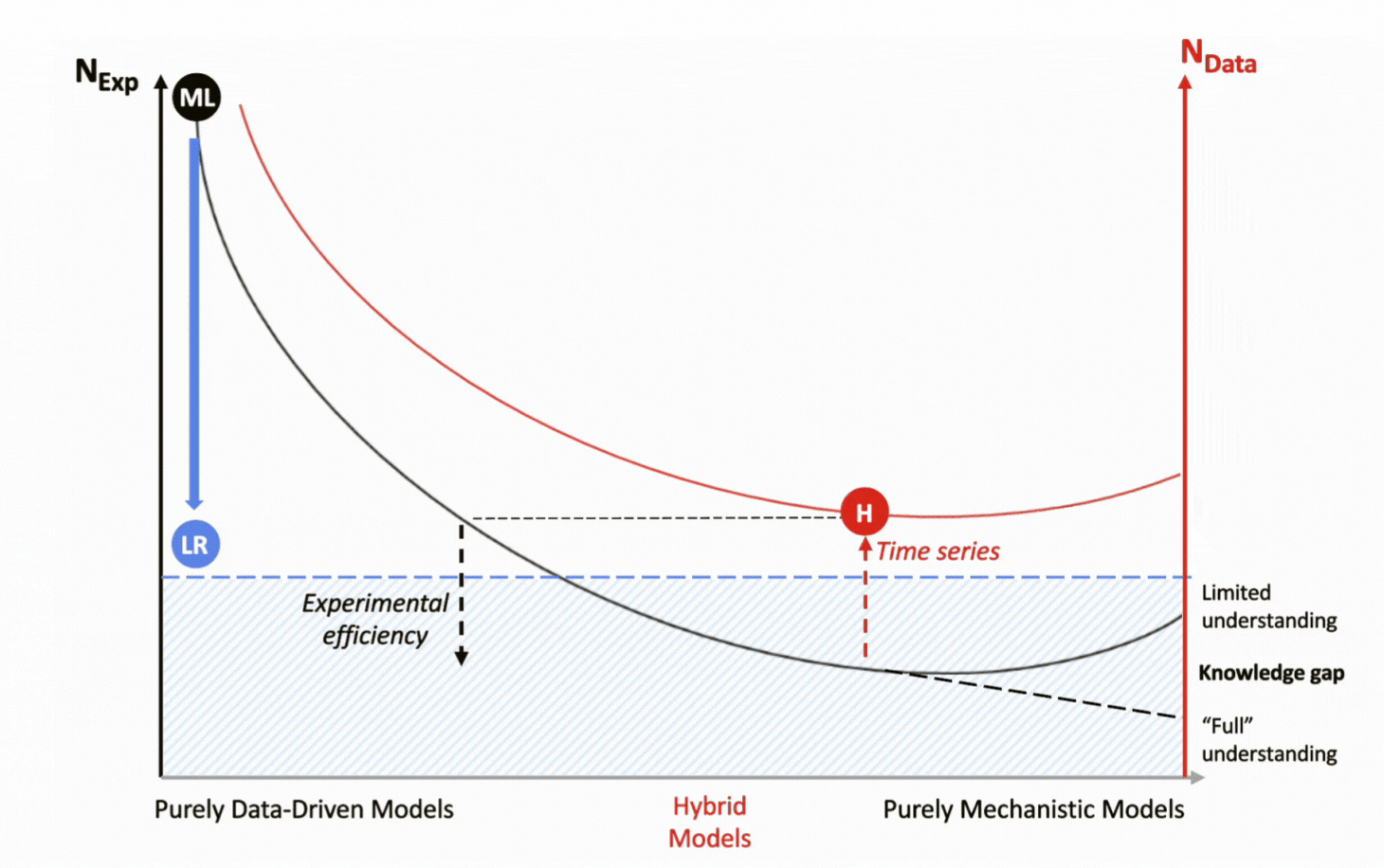
For over a decade, DataHow has pioneered the application of hybrid models to bioprocessing. By integrating a mechanistic backbone with a data-driven machine learning layer, they combine the strengths of both approaches, delivering accuracy while remaining highly data-efficient.
The two elements of the hybrid model work in a complementary fashion. The mechanistic backbone transports the domain’s scientific knowledge to describe well-established process dynamics, while the machine learning layer fills the gaps by capturing unknown, complex, and nonlinear behaviors. Together, they create models that are both interpretable and predictive, even under data-constrained conditions.
With their mechanistic backbone, hybrid models capture full process dynamics by leveraging entire time-course trajectories rather than relying on static end-points as statistical regression models do. This approach not only reveals how the process evolves but also amplifies the informational value of each experiment, further reducing the need for new data generation through experimentation.
![]() Pure machine learning models require too much experimental data to be viable in R&D;
Pure machine learning models require too much experimental data to be viable in R&D;
![]() Linear regression models make compromises on model quality and process knowledge to focus on main effects with less data;
Linear regression models make compromises on model quality and process knowledge to focus on main effects with less data;
![]() LR Models only focus on endpoints: 1 experiment = 1 data point;
LR Models only focus on endpoints: 1 experiment = 1 data point;
![]() Hybrid introduces knowledge mechanistically, reducing data dependency and captures time series data for each experiment:
Hybrid introduces knowledge mechanistically, reducing data dependency and captures time series data for each experiment:
1 experiment = multiple data points;
![]() Hybrid supports experimental efficiency.
Hybrid supports experimental efficiency.
Approaching Experimental Design with Hybrid Technology
Unlike linear regression, hybrid models are capable of capturing the complex, non-linear, and combinatorial interactions that naturally arise in bioprocesses. This ability fundamentally changes how experimental campaigns should be designed.
Instead of limiting experiments to corner or center points, space-filling designs such as Latin hypercube sampling distribute them uniformly across the design space. This generates richer datasets for hybrid models, enabling more accurate insights with fewer, strategically placed experiments tailored to time and budget constraints. This design approach, optimal for hybrid models, enables efficient initial data generation, reducing the total number of experiments needed to learn and optimize process performance without sacrificing any design factors to ease the experimental burden.
DataHow has further advanced this approach by creating a dedicated development methodology to complement its hybrid modeling technology. This methodology integrates space-filling designs into short, iterative experimental loops that accelerate time to insight and enable developers to plan experiments deliberately, guided by the latest findings.
This marks a paradigm shift in bioprocess development, both operationally and in outcomes. Rather than running disconnected, large-scale campaigns tailored to linear analytics, development shifts toward shorter, interlinked experimental loops, guided by insight and powered by technology capable of handling the inherent non-linearity of bioprocesses.
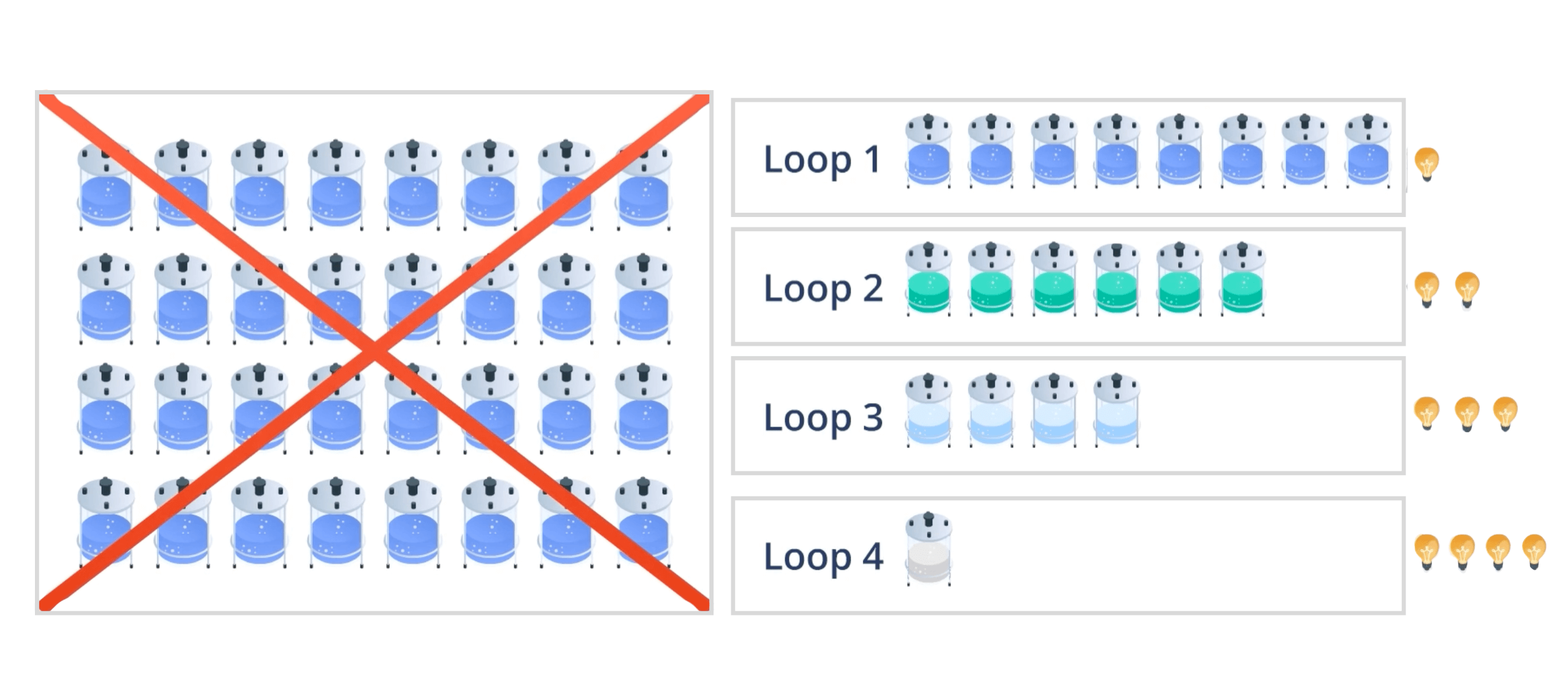
Transfer Learning: Using Historical Knowledge
Many organizations repeatedly develop analogous processes, representing months and years of accumulated experimental data, process information, and domain knowledge. Yet despite this extensive knowledge base, new development projects are typically initiated as if no prior learning existed, commencing with a de novo Design of Experiments (DoE) campaign that treats the system as an entirely unexplored design space. The same can be said for scale-up, where small-scale data is practically abandoned as large-scale analysis begins.
Machine learning methods offer powerful support in this regard, transforming historical data from a sunk cost into a valuable development asset. DataHow’s transfer learning technology draws on accumulated historical datasets to extract insights from relevant, similar processes and transfer them to current projects. Similarly, scale-up efforts can leverage small-scale data, using large-scale runs only where knowledge is missing or model uncertainty is high. Transfer learning, therefore, transforms existing data into accessible knowledge, overcoming a key limitation of traditional DoE models that lack the ability to transfer knowledge across projects.
DataHow’s approach treats experiments as a scarce resource, an input to be used intentionally and only where existing knowledge, either introduced through the mechanistic backbone or available data, is insufficient. As your database of knowledge expands, so does the scope for efficiency in your next project. Imagine the terabytes of efficiency gains sitting in the servers of Big Pharma.

The DataHow Digital Development Framework
DataHow has been developing the core hybrid modeling technologies for bioprocessing for over a decade. As the technologies have matured, so too has our understanding of how to best deploy them in development. The result is technologically innovative and scientifically rigorous, while being operationally pragmatic and efficient. Importantly, it is simple to use.
DataHow’s digital framework is built around iterative development loops:
-
Transfer learning or space-filling designs where no prior knowledge exists
-
Insight-informed experimental design toward process objectives
-
Execution of experiments in the lab (and direct data transfer through connectors)
-
Data preparation
-
Model training and evaluation
-
Insight generation and goal assessment
-
Progress to the next development stage or further iteration with targeted experiments
The framework and development loops are operationalized with DataHow’s bioprocess intelligence platform, DataHowLab, where technology, method, and user experience are merged to support operational objectives. It enables users to navigate through each step of the loop, iteratively building four key digital assets:

| Data
Generated from experimental sprints or transferred from historical projects. |
Models
Advanced hybrid models learn process dynamics from new and relevant historical process data. |
Insights
Models generate insight to support decision-making and further optimization of the process. |
Recipes
Insights inform next experiments and the convergence towards an optimal recipe. |
Dynamic Process Knowledge Deployment as Digital Twins
As the process moves toward commercial manufacturing, these assets form a consolidated body of process knowledge, complete with a digital trail spanning from early to late-stage development. This offers a potential step change in digital bioprocessing: technology transfer moves beyond static reporting packages to dynamic digital assets that encode the full continuum of development knowledge. Comprehensive datasets, process insights, complete process models, and optimized recipes are all digitally integrated and ready for use in manufacturing.
Trained across all lifecycle stages and exposed to broad process variability, these models encode the ability to optimize and correct toward defined performance objectives. Integrated into manufacturing monitoring and control systems, they continuously process real-time data to generate predictive forecasts and flag potential deviations. Human-in-the-loop frameworks allow operators to test remediation strategies virtually before applying them on the plant floor. As confidence in these systems grows, the path toward fully automated, closed-loop process control becomes increasingly realistic.
A Technology for Process Scientists
Hybrid modeling is increasingly recognized as the future of bioprocess analytics and a key enabler of efficient digital bioprocessing. Yet for many organizations and service providers, the emphasis remains on the technology itself, focusing on building the engine rather than the entire car. As a result, these solutions often remain confined to technologists and data scientists rather than being broadly adopted across development teams.
However, for technology to be fully embraced and operationally impactful, it must be placed in the hands of those who use it daily: the scientists and engineers on the shop floor. DataHowLab has been built with this philosophy at its core, embedding advanced technology into intuitive workflows and guided tasks that help users achieve their objectives efficiently and effectively. The platform positions models as the means to an end rather than the goal, the enabler rather than the star of the show. Model coding is replaced by model libraries and workflows, and insight and knowledge are created with minimal expertise in data science.
This platform aims to reduce the distance between data generators (the scientists) and insight enablers (data analytics and modeling), empowering teams with their data without the data-science disconnect. Yes, a degree of upskilling is required to be comfortable with analytical approaches and techniques, but this is a natural development in an increasingly data-fueled domain.
New Tools and Technology: A New Way Forward
Bioprocess development has always advanced through the tools available to it, and DoE has been instrumental in bringing rigor and structure to the field. Today, hybrid modeling builds on that foundation, offering a way to handle the greater complexity of modern processes while improving operational efficiency. Platforms like DataHowLab make this evolution practical, translating powerful modeling approaches into accessible workflows for scientists and engineers. As with every major step forward in the field, new tools and technologies demand new ways of working. If the industry is committed to its vision of Pharma 4.0 and smart digital bioprocessing, it must embrace change.
Ready to make the change? Speak to the DataHow team about how new technologies and methods could transform your operation.